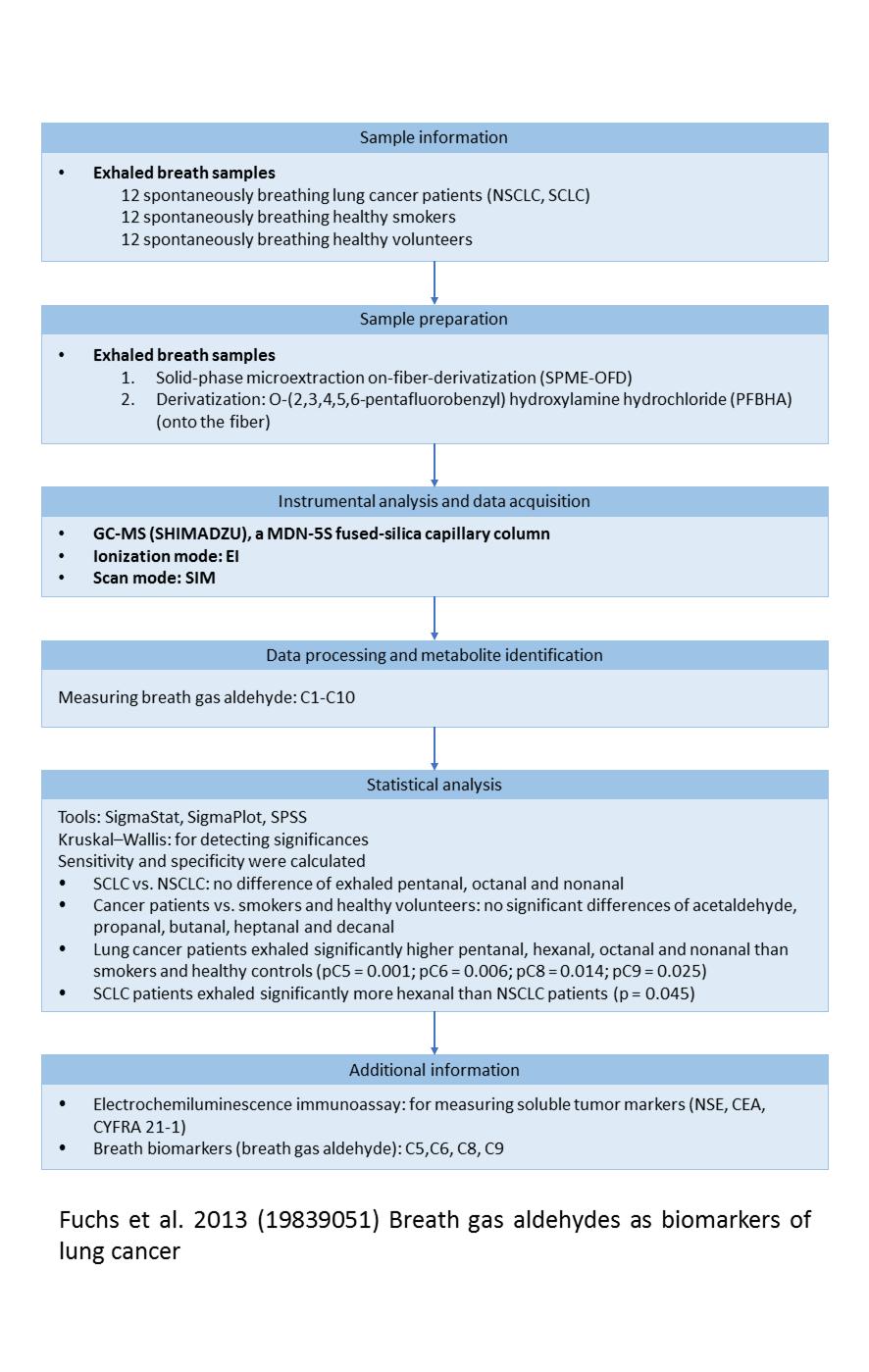
Showing paper detailed information
Citation Information
Mol Biosyst. 2013 Sep;9(9):2370-8. doi: 10.1039/c3mb70138g.
Exploratory investigation of plasma metabolomics in human lung adenocarcinoma.
Wen T, Gao L, Wen Z, Wu C, Tan CS, Toh WZ, Ong CN.
Analytical methods | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Chromatography | GC | LC | |
| Ion source | EI | ESI | |
| Positive/Negative mode | – | – | |
| Mass analyzer | – | Q-TOF | |
| Identification level | – | MS/MS | |
Sample information | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Country | China | China | |
| Specimen | plasma | plasma | |
| Marker function | diagnosis | diagnosis | |
| Participants(Case) | Cancer type | adenocarcinoma | adenocarcinoma |
| Stage | I | I | |
| Number | 31 | 31 | |
| Gender (M,F) | 15, 16 | 15, 16 | |
| Mean age (range) (M,F) | median: 63 (40-81) | median: 63 (40-81) | |
| Smoking status | smoker, non-smoker | smoker, non-smoker | |
| Participants(Control) | Type | healthy | healthy |
| Number | 28 | 28 | |
| Gender (M,F) | 20, 8 | 20, 8 | |
| Mean age (range) (M,F) | median: 37 (29-50) | median: 37 (29-50) | |
| Smoking status | smoker, non-smoker | smoker, non-smoker | |
Data processing and metabolite identification | |
|---|---|
| Data processing software | MassHunter, Mass Profiler Professional software (Agilent) |
| Database search | NIST 08, HMDB, METLIN, LIPID MAPS |
Statistical analysis | |
|---|---|
| Differential analysis method | Mann–Whitney–Wilcoxon test, OPLS-DA |
| Classification method | ROC curve analysis |
| Survival analysis method | – |
Lung cancer-related metabolites identified in the paper
| Metabolite | Author-emphasized biomarkers | Mean concentration (case) | Mean concentration (control) | Fold change (case/control) | P-value | FDR | VIP |
|---|---|---|---|---|---|---|---|
| 5-hydroxytryptophan | – | – | 0.37 | 7.72e-03 | – | 1.19 | |
| amylose | – | – | – | 709.18 | 4.09e-11 | – | 1.34 |
| bilirubin | – | – | – | 290.02 | 3.15e-10 | – | 1.23 |
| maltitol | – | – | – | 61.82 | 4.35e-09 | – | 1.25 |
| N-palmitoleoyl ethanolamide | – | – | – | 19.03 | 5.14e-09 | – | 1.03 |
| oleic acid | – | – | – | 15.67 | 2.45e-10 | – | 1.28 |
| linoleic acid | – | – | – | 7.46 | 4.41e-10 | – | 1.25 |
| ubiquinone | – | – | – | 5.62 | 1.15e-09 | – | 1.29 |
| ubiquinol | – | – | – | 5.54 | 1.53e-09 | – | 1.23 |
| palmitic acid | – | – | – | 4.26 | 1.79e-08 | – | 1.16 |
| glutamine | – | – | – | 0.44 | 6.52e-03 | – | 1.40 |
| alanine | – | – | – | 0.38 | 7.12e-04 | – | 1.39 |
| 3-hydroxybutyric acid | – | – | – | 0.36 | 1.46e-03 | – | 1.57 |
| deoxycholic acid glycine conjugate | – | – | – | 0.35 | 4.76e-04 | – | 1.05 |
| PE(38:6) | – | – | – | 0.35 | 1.48e-04 | – | 1.18 |
| glycine | – | – | – | 0.33 | 3.19e-05 | – | 1.51 |
| pipecolic acid | – | – | – | 0.33 | 1.19e-05 | – | 1.16 |
| PE(36:2) | – | – | – | 0.32 | 9.44e-07 | – | 1.12 |
| threonine | – | – | – | 0.30 | 4.50e-03 | – | 1.44 |
| PE(34:2) | – | – | – | 0.30 | 9.44e-07 | – | 1.12 |
| uric acid | – | – | – | 0.27 | 1.32e-05 | – | 1.26 |
| pregnenolone sulfate | – | – | – | 0.19 | 1.16e-05 | – | 1.23 |
| N(6)-Methyllysine | – | – | – | 0.15 | 4.23e-04 | – | 1.17 |
| proline | – | – | – | 0.15 | 5.49e-08 | – | 1.03 |
| PC(34:4) | – | – | – | 0.13 | 5.27e-07 | – | 1.22 |
| L-arginine | – | – | – | 0.13 | 1.79e-07 | – | 1.13 |
| PE(36:4) | – | – | – | 0.13 | 1.27e-07 | – | 1.31 |
| androsterone sulfate | – | – | – | 0.13 | 3.53e-09 | – | 1.11 |
| PS(38:4) | – | – | – | 0.10 | 1.52e-05 | – | 1.17 |
| PE(38:4) | – | – | – | 0.10 | 9.52e-10 | – | 1.23 |
| testosterone sulfate | – | – | – | 0.10 | 4.86e-10 | – | 1.22 |
| ceramides (42:0) | – | – | – | 0.08 | 8.93e-07 | – | 1.08 |
| PE(36:1) | – | – | – | 0.06 | 1.44e-10 | – | 1.36 |
| PE(40:5) | – | – | – | 0.01 | 8.09e-11 | – | 1.30 |
| PE(40:4) | – | – | – | 9.55e-03 | 3.26e-08 | – | 1.07 |
| 3,4,5-trimethoxycinnamic acid | – | – | – | – | 3.16e-12 | – | 1.62 |
| 5-methoxytryptophan | – | – | – | – | 3.16e-12 | – | 1.61 |
| Metabolite | Author-emphasized biomarkers | Cutoff value | AUROC (95%CI) | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| 5-hydroxytryptophan | – | – | 0.7 | – | – | – |
| amylose | – | – | 0.98 | – | – | – |
| bilirubin | – | – | 0.96 | – | – | – |
| maltitol | – | – | 0.93 | – | – | – |
| N-palmitoleoyl ethanolamide | – | – | 0.93 | – | – | – |
| oleic acid | – | – | 0.98 | – | – | – |
| linoleic acid | – | – | 0.97 | – | – | – |
| ubiquinone | – | – | 0.96 | – | – | – |
| ubiquinol | – | – | 0.96 | – | – | – |
| palmitic acid | – | – | 0.93 | – | – | – |
| glutamine | – | – | 0.71 | – | – | – |
| alanine | – | – | 0.76 | – | – | – |
| 3-hydroxybutyric acid | – | – | 0.74 | – | – | – |
| deoxycholic acid glycine conjugate | – | – | 0.76 | – | – | – |
| PE(38:6) | – | – | 0.79 | – | – | – |
| glycine | – | – | 0.82 | – | – | – |
| pipecolic acid | – | – | 0.83 | – | – | – |
| PE(36:2) | – | – | 0.87 | – | – | – |
| threonine | – | – | 0.71 | – | – | – |
| PE(34:2) | – | – | 0.87 | – | – | – |
| uric acid | – | – | 0.83 | – | – | – |
| pregnenolone sulfate | – | – | 0.83 | – | – | – |
| N(6)-Methyllysine | – | – | 0.75 | – | – | – |
| proline | – | – | 0.9 | – | – | – |
| PC(34:4) | – | – | 0.85 | – | – | – |
| L-arginine | – | – | 0.89 | – | – | – |
| PE(36:4) | – | – | 0.9 | – | – | – |
| androsterone sulfate | – | – | 0.95 | – | – | – |
| PS(38:4) | – | – | 0.81 | – | – | – |
| PE(38:4) | – | – | 0.96 | – | – | – |
| testosterone sulfate | – | – | 0.97 | – | – | – |
| ceramides (42:0) | – | – | 0.86 | – | – | – |
| PE(36:1) | – | – | 0.97 | – | – | – |
| PE(40:5) | – | – | 0.98 | – | – | – |
| PE(40:4) | – | – | 0.9 | – | – | – |
| 3,4,5-trimethoxycinnamic acid | – | – | 1 | – | – | – |
| 5-methoxytryptophan | – | – | 1 | – | – | – |
Paper graphical summary