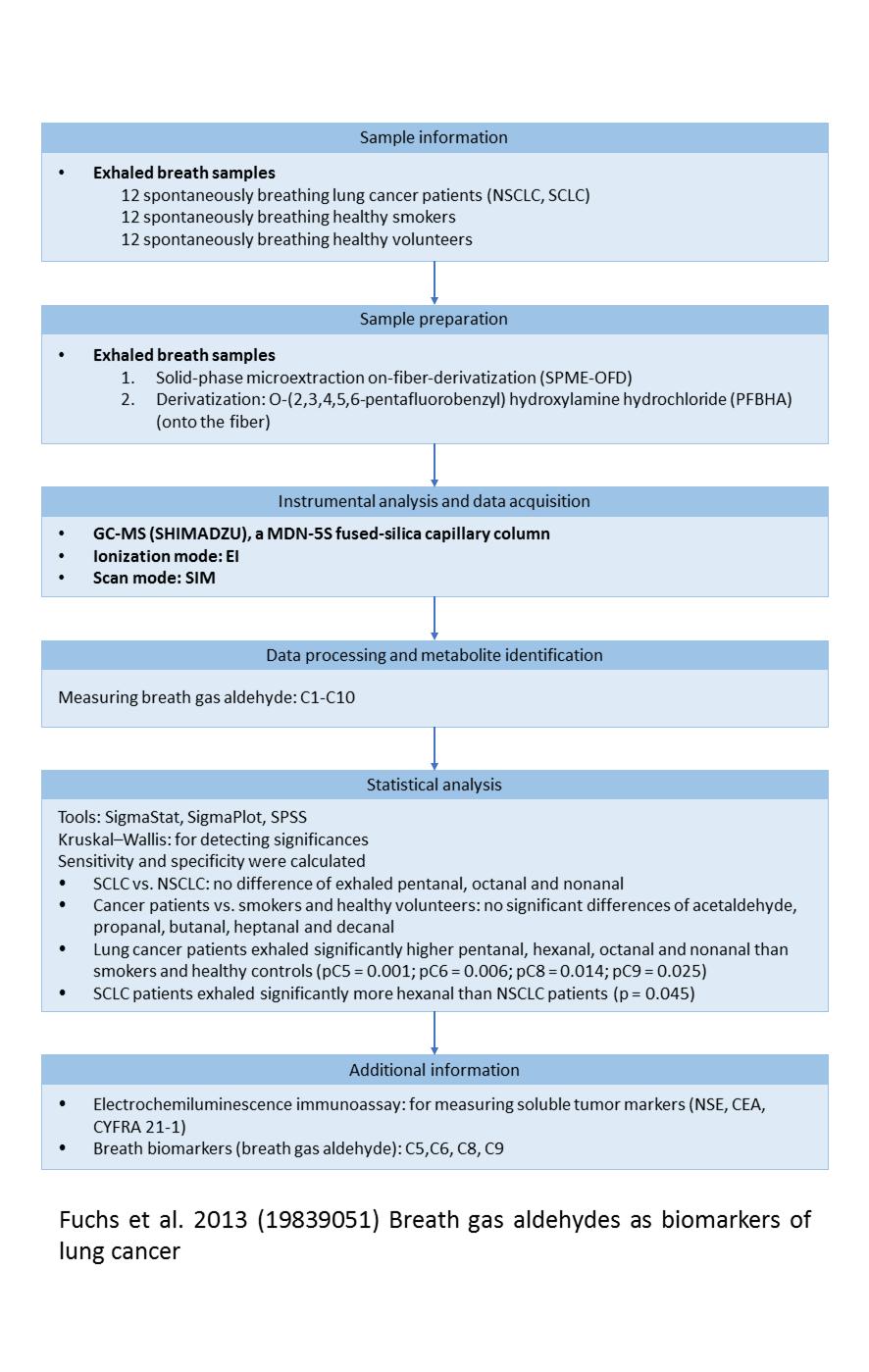
Showing paper detailed information
Citation Information
J Breath Res. 2014 Jun;8(2):027111. doi: 10.1088/1752-7155/8/2/027111. Epub 2014 May 27.
Comparative analyses of volatile organic compounds (VOCs) from patients, tumors and transformed cell lines for the validation of lung cancer-derived breath markers.
Filipiak W, Filipiak A, Sponring A, Schmid T, Zelger B, Ager C, Klodzinska E, Denz H, Pizzini A, Lucciarini P, Jamnig H, Troppmair J, Amann A.
Analytical methods | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Chromatography | GC | GC | |
| Ion source | – | – | |
| Positive/Negative mode | – | – | |
| Mass analyzer | – | – | |
| Identification level | – | – | |
Sample information | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Country | Austria | Austria | |
| Specimen | exhaled breath | tissue | |
| Marker function | diagnosis | diagnosis | |
| Participants(Case) | Cancer type | SCLC, squamous cell carcinoma, adenocarcinoma, large cell carcinoma, mesothelioma, carcinoid | NSCLC, SCLC |
| Stage | – | I, II, III, IV | |
| Number | 36 | 14 | |
| Gender (M,F) | 25, 11 | 6, 8 | |
| Mean age (range) (M,F) | 63 ± 7 | 67.7, 63.4 | |
| Smoking status | former, non-smoker | smoker, non-smoker | |
| Participants(Control) | Type | healthy | tumor vs. adjacent normal tissue |
| Number | 28 | 13 | |
| Gender (M,F) | 12, 16 | 12, 16 | |
| Mean age (range) (M,F) | 52 ± 17 | 52 ± 17 | |
| Smoking status | former, non-smokers | former, non-smokers | |
Data processing and metabolite identification | |
|---|---|
| Data processing software | – |
| Database search | NIST 2005 |
Statistical analysis | |
|---|---|
| Differential analysis method | Kruskal–Wallis test, Spearman |
| Classification method | Kruskal–Wallis test, Spearman |
| Survival analysis method | – |
Lung cancer-related metabolites identified in the paper
| Metabolite | Author-emphasized biomarkers | Mean concentration (case) | Mean concentration (control) | Fold change (case/control) | P-value | FDR | VIP |
|---|---|---|---|---|---|---|---|
| 1(R)-a-pinene | – | – | – | – | – | – | – |
| 1(R)-a-pinene | – | – | – | – | – | – | – |
| 2,3,4-trimethylpentane | – | – | – | – | – | – | – |
| 2,3,4-trimethylpentane | – | – | – | – | – | – | – |
| 2,4-dimethyl-1-heptene | – | – | – | – | – | – | – |
| 2,4-dimethyl-1-heptene | – | – | – | – | – | – | – |
| 2,4-dimethylheptane | – | – | – | – | – | – | – |
| 2,4-dimethylheptane | – | – | – | – | – | – | – |
| 2-butanon | – | – | – | – | – | – | – |
| 2-butanon | – | – | – | – | – | – | – |
| 2-butenal | – | – | – | – | – | – | – |
| 2-butenal | – | – | – | – | – | – | – |
| 2-hexanone | – | – | – | – | – | – | – |
| 2-hexanone | – | – | – | – | – | – | – |
| 2-methyl-1-pentene | – | – | – | – | – | – | – |
| 2-methyl-1-pentene | – | – | – | – | – | – | – |
| 2-methylbutanal | – | – | – | – | – | – | – |
| 2-methylbutanal | – | – | – | – | – | – | – |
| 2-methylpentane | – | – | – | – | – | – | – |
| 2-methylpentane | – | – | – | – | – | – | – |
| 2-methylpropanal | – | – | – | – | – | – | – |
| 2-methylpropanal | – | – | – | – | – | – | – |
| 2-pentanone | – | – | – | – | – | – | – |
| 2-pentanone | – | – | – | – | – | – | – |
| 2-propanol | – | – | – | – | – | – | – |
| 2-propanol | – | – | – | – | – | – | – |
| 2-propenal | – | – | – | – | – | – | – |
| 2-propenal | – | – | – | – | – | – | – |
| 3-methylbutanal | – | – | – | – | – | – | – |
| 3-methylbutanal | – | – | – | – | – | – | – |
| 3-methylhexane | – | – | – | – | – | – | – |
| 3-methylhexane | – | – | – | – | – | – | – |
| 4-methylheptane | – | – | – | – | – | – | – |
| 4-methylheptane | – | – | – | – | – | – | – |
| 4-methyloctane | – | – | – | – | – | – | – |
| 4-methyloctane | – | – | – | – | – | – | – |
| 6-methyl-5-heptene-2-one | – | – | – | – | – | – | – |
| 6-methyl-5-heptene-2-one | – | – | – | – | – | – | – |
| acetaldehyde | – | – | – | – | – | – | – |
| acetaldehyde | – | – | – | – | – | – | – |
| acetic acid | – | – | – | – | – | – | – |
| acetic acid | – | – | – | – | – | – | – |
| acetone | – | – | – | – | – | – | – |
| acetone | – | – | – | – | – | – | – |
| acetonitrile | – | – | – | – | – | – | – |
| acetonitrile | – | – | – | – | – | – | – |
| benzaldehyde | – | – | – | – | – | – | – |
| benzaldehyde | – | – | – | – | – | – | – |
| butanedione | – | – | – | – | – | – | – |
| butanedione | – | – | – | – | – | – | – |
| decanal | – | – | – | – | – | – | – |
| decanal | – | – | – | – | – | – | – |
| ethanol | V | – | – | – | – | – | – |
| ethanol | – | – | – | – | – | – | – |
| hexanal | – | – | – | – | – | – | – |
| hexanal | – | – | – | – | – | – | – |
| hexane | – | – | – | – | – | – | – |
| hexane | – | – | – | – | – | – | – |
| methanol | – | – | – | – | – | – | – |
| methanol | – | – | – | – | – | – | – |
| methyl acetate | – | – | – | – | – | – | – |
| methyl acetate | – | – | – | – | – | – | – |
| N-dodecan | – | – | – | – | – | – | – |
| N-dodecan | – | – | – | – | – | – | – |
| nonane | – | – | – | – | – | – | – |
| nonane | – | – | – | – | – | – | – |
| octanal | V | – | – | – | – | – | – |
| octanal | – | – | – | – | – | – | – |
| octane | – | – | – | – | – | – | – |
| octane | – | – | – | – | – | – | – |
| propanal | – | – | – | – | – | – | – |
| propanal | – | – | – | – | – | – | – |
| propene | – | – | – | – | – | – | – |
| propene | – | – | – | – | – | – | – |
| pyridine | – | – | – | – | – | – | – |
| pyridine | – | – | – | – | – | – | – |
| pyrrole | – | – | – | – | – | – | – |
| pyrrole | – | – | – | – | – | – | – |
| Metabolite | Author-emphasized biomarkers | Cutoff value | AUROC (95%CI) | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| 1(R)-a-pinene | – | – | – | – | – | – |
| 1(R)-a-pinene | – | – | – | – | – | – |
| 2,3,4-trimethylpentane | – | – | – | – | – | – |
| 2,3,4-trimethylpentane | – | – | – | – | – | – |
| 2,4-dimethyl-1-heptene | – | – | – | – | – | – |
| 2,4-dimethyl-1-heptene | – | – | – | – | – | – |
| 2,4-dimethylheptane | – | – | – | – | – | – |
| 2,4-dimethylheptane | – | – | – | – | – | – |
| 2-butanon | – | – | – | – | – | – |
| 2-butanon | – | – | – | – | – | – |
| 2-butenal | – | – | – | – | – | – |
| 2-butenal | – | – | – | – | – | – |
| 2-hexanone | – | – | – | – | – | – |
| 2-hexanone | – | – | – | – | – | – |
| 2-methyl-1-pentene | – | – | – | – | – | – |
| 2-methyl-1-pentene | – | – | – | – | – | – |
| 2-methylbutanal | – | – | – | – | – | – |
| 2-methylbutanal | – | – | – | – | – | – |
| 2-methylpentane | – | – | – | – | – | – |
| 2-methylpentane | – | – | – | – | – | – |
| 2-methylpropanal | – | – | – | – | – | – |
| 2-methylpropanal | – | – | – | – | – | – |
| 2-pentanone | – | – | – | – | – | – |
| 2-pentanone | – | – | – | – | – | – |
| 2-propanol | – | – | – | – | – | – |
| 2-propanol | – | – | – | – | – | – |
| 2-propenal | – | – | – | – | – | – |
| 2-propenal | – | – | – | – | – | – |
| 3-methylbutanal | – | – | – | – | – | – |
| 3-methylbutanal | – | – | – | – | – | – |
| 3-methylhexane | – | – | – | – | – | – |
| 3-methylhexane | – | – | – | – | – | – |
| 4-methylheptane | – | – | – | – | – | – |
| 4-methylheptane | – | – | – | – | – | – |
| 4-methyloctane | – | – | – | – | – | – |
| 4-methyloctane | – | – | – | – | – | – |
| 6-methyl-5-heptene-2-one | – | – | – | – | – | – |
| 6-methyl-5-heptene-2-one | – | – | – | – | – | – |
| acetaldehyde | – | – | – | – | – | – |
| acetaldehyde | – | – | – | – | – | – |
| acetic acid | – | – | – | – | – | – |
| acetic acid | – | – | – | – | – | – |
| acetone | – | – | – | – | – | – |
| acetone | – | – | – | – | – | – |
| acetonitrile | – | – | – | – | – | – |
| acetonitrile | – | – | – | – | – | – |
| benzaldehyde | – | – | – | – | – | – |
| benzaldehyde | – | – | – | – | – | – |
| butanedione | – | – | – | – | – | – |
| butanedione | – | – | – | – | – | – |
| decanal | – | – | – | – | – | – |
| decanal | – | – | – | – | – | – |
| ethanol | V | – | – | – | – | – |
| ethanol | – | – | – | – | – | – |
| hexanal | – | – | – | – | – | – |
| hexanal | – | – | – | – | – | – |
| hexane | – | – | – | – | – | – |
| hexane | – | – | – | – | – | – |
| methanol | – | – | – | – | – | – |
| methanol | – | – | – | – | – | – |
| methyl acetate | – | – | – | – | – | – |
| methyl acetate | – | – | – | – | – | – |
| N-dodecan | – | – | – | – | – | – |
| N-dodecan | – | – | – | – | – | – |
| nonane | – | – | – | – | – | – |
| nonane | – | – | – | – | – | – |
| octanal | V | – | – | – | – | – |
| octanal | – | – | – | – | – | – |
| octane | – | – | – | – | – | – |
| octane | – | – | – | – | – | – |
| propanal | – | – | – | – | – | – |
| propanal | – | – | – | – | – | – |
| propene | – | – | – | – | – | – |
| propene | – | – | – | – | – | – |
| pyridine | – | – | – | – | – | – |
| pyridine | – | – | – | – | – | – |
| pyrrole | – | – | – | – | – | – |
| pyrrole | – | – | – | – | – | – |
Paper graphical summary