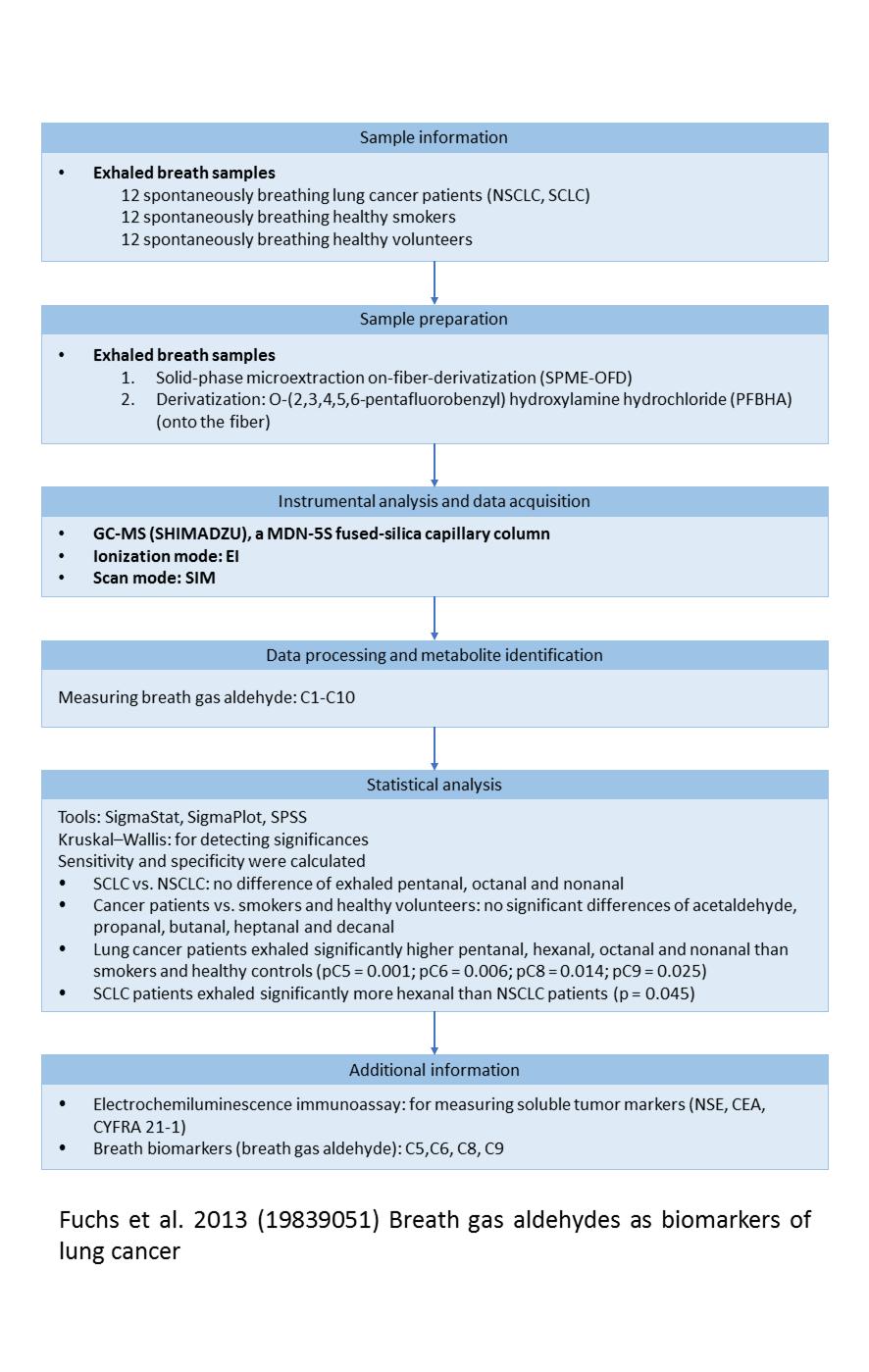
Showing paper detailed information
Citation Information
J Proteome Res. 2020 Sep 4;19(9):3750-3760. doi: 10.1021/acs.jproteome.0c00285. Epub 2020 Jul 31.
Liquid Chromatography-Mass Spectrometry-Based Tissue Metabolic Profiling Reveals Major Metabolic Pathway Alterations and Potential Biomarkers of Lung Cancer.
You L, Fan Y, Liu X, Shao S, Guo L, Noreldeen HAA, Li Z, Ouyang Y, Li E, Pan X, Liu T, Tian X, Ye F, Li X, Xu G.
Analytical methods | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Chromatography | LC | LC | |
| Ion source | ESI | ESI | |
| Positive/Negative mode | both | both | |
| Mass analyzer | Q-TOF | Q-TOF | |
| Identification level | MS/MS | MS/MS | |
Sample information | |||
|---|---|---|---|
| Comparative study | #1 | #2 | |
| Country | China | China | |
| Specimen | tissue | tissue | |
| Marker function | diagnosis | diagnosis | |
| Participants(Case) | Cancer type | AC | NSCLC |
| Stage | I, II, III | I, II, III | |
| Number | 45 | 85 | |
| Gender (M,F) | – | 51, 34 | |
| Mean age (range) (M,F) | – | 59 ± 12 | |
| Smoking status | – | – | |
| Participants(Control) | Type | SCC | benign lung tissues |
| Number | 18 | 85 | |
| Gender (M,F) | – | 51, 34 | |
| Mean age (range) (M,F) | – | 59 ± 12 | |
| Smoking status | – | – | |
Data processing and metabolite identification | |
|---|---|
| Data processing software | MarkerView workstation, MultiQuant 3.0.3 |
| Database search | OSI-SMMS |
Statistical analysis | |
|---|---|
| Differential analysis method | PCA, PLS-DA, non-parametric test |
| Classification method | – |
| Survival analysis method | – |
Lung cancer-related metabolites identified in the paper
| Metabolite | Author-emphasized biomarkers | Mean concentration (case) | Mean concentration (control) | Fold change (case/control) | P-value | FDR | VIP |
|---|---|---|---|---|---|---|---|
| creatine | – | – | – | 0.34 | <0.001 | <0.001 | – |
| 6-phosphonoglucono-D-lactone | – | – | – | 2.00 | 1.00e-03 | 0.03 | – |
| 2-hydroxy-6-aminopurine | – | – | – | 2.42 | <0.001 | <0.001 | – |
| 8-hydroxy-2-deoxyguanosine | – | – | – | 2.44 | 1.00e-03 | 0.03 | – |
| guanosine | – | – | – | 2.27 | 1.00e-03 | 0.03 | – |
| myo-inositol | – | – | – | 2.03 | <0.001 | <0.001 | – |
| Cer(d18:0/16:0)-2 | – | – | – | 0.29 | <0.001 | <0.001 | – |
| LPC 15:0 sn-1 | – | – | – | 2.87 | <0.001 | <0.001 | – |
| LPC 17:0 sn-1 | – | – | – | 2.18 | 2.00e-03 | 0.04 | – |
| LPE 16:0 sn-1 | – | – | – | 3.05 | 2.00e-03 | 0.04 | – |
| LPE 18:1 sn-1 | – | – | – | 2.73 | 1.00e-03 | 0.03 | – |
| PC 36:0 | – | – | – | 0.54 | 2.00e-03 | 0.04 | – |
| PC 38:1 | – | – | – | 0.38 | <0.001 | <0.001 | – |
| PC O 36:1 | – | – | – | 0.55 | 2.00e-03 | 0.04 | – |
| SM 35:2 | – | – | – | 1.24 | 1.00e-03 | 0.03 | – |
| 6-phosphogluconic acid | – | – | – | 1.65 | 1.00e-03 | – | – |
| citric acid | – | – | – | 1.66 | 1.00e-03 | – | – |
| lactic acid | – | – | – | 0.66 | 1.00e-03 | – | – |
| glyceraldehyde-3-phosphate | – | – | – | 1.24 | 0.04 | – | – |
| D-glucose 6-phosphate | – | – | – | 1.85 | 1.00e-03 | – | – |
| D-Sedoheptuiose 7-phosphate | – | – | – | 1.84 | 2.00e-03 | – | – |
| D-Erythrose 4-phosphate | – | – | – | 1.78 | 2.00e-03 | – | – |
| 5-deoxy-5'-methylthioadenosine | – | – | – | 0.49 | 4.00e-03 | – | – |
| SAH | – | – | – | 0.64 | 0.03 | – | – |
| xanthine | – | – | – | 0.47 | <0.001 | – | – |
| creatine | – | – | – | 0.60 | 0.03 | – | – |
| glutathione | – | – | – | 0.22 | 0.02 | – | – |
| oxidized glutathione | – | – | – | 0.50 | 0.04 | – | – |
| carnitine C4:0 | – | – | – | 0.75 | 0.02 | – | – |
| carnitine C18:1 | – | – | – | 0.81 | 0.04 | – | – |
| FFA C20:2 | – | – | – | 0.75 | 0.05 | – | – |
| FFA C20:3 | – | – | – | 0.71 | 0.03 | – | – |
| Eicosatrienoic acid methyl ester | – | – | – | 0.91 | 6.00e-03 | – | – |
| LPC 15:0 sn-1 | – | – | – | 0.40 | 6.00e-03 | – | – |
| LPC 15:0 sn-2 | – | – | – | 0.64 | 0.04 | – | – |
| LPC 18:1 sn-2 | – | – | – | 0.73 | 0.04 | – | – |
| LPC 22:0 sn-2 | – | – | – | 1.93 | 8.00e-03 | – | – |
| LPC 22:4 sn-2 | – | – | – | 0.74 | 0.03 | – | – |
| LPE 16:1 sn-1 | – | – | – | 0.55 | 0.01 | – | – |
| LPE 16:1 sn-2 | – | – | – | 0.58 | 0.02 | – | – |
| LPE 18:0 sn-1 | – | – | – | 0.51 | 0.04 | – | – |
| LPE 18:1 sn-2 | – | – | – | 0.72 | 8.00e-03 | – | – |
| LPE 20:1 sn-1 | – | – | – | 0.30 | 1.00e-03 | – | – |
| LPE 22:4 sn-2 | – | – | – | 0.80 | 0.03 | – | – |
| LPE 22:6 sn-2 | – | – | – | 0.75 | 6.00e-03 | – | – |
| PC 32:0 | – | – | – | 1.22 | 0.01 | – | – |
| PC 33:1 | – | – | – | 0.63 | 1.00e-03 | – | – |
| PC 33:2 | – | – | – | 0.70 | 3.00e-03 | – | – |
| PC 35:2 | – | – | – | 0.62 | <0.001 | – | – |
| PC 35:3 | – | – | – | 0.67 | <0.001 | – | – |
| PC O 34:2-2 | – | – | – | 0.67 | 1.00e-03 | – | – |
| PE O 36:3 | – | – | – | 0.64 | 1.00e-02 | – | – |
| PE 32:1 | – | – | – | 0.56 | 2.00e-03 | – | – |
| PE 38:1 | – | – | – | 0.61 | 1.00e-03 | – | – |
| PE 38:4 | – | – | – | 0.76 | 7.00e-03 | – | – |
| PE 38:6 | – | – | – | 0.80 | 0.05 | – | – |
| PE O 34:3 | – | – | – | 0.44 | 5.00e-03 | – | – |
| SM 36:2 | – | – | – | 1.23 | 0.03 | – | – |
| sphinganine | – | – | – | 1.65 | 0.02 | – | – |
| sphingosine | – | – | – | 1.40 | 0.02 | – | – |
| kynurenine | – | – | – | 0.62 | 0.03 | – | – |
| oxypurinol | – | – | – | 0.43 | <0.001 | – | – |
| pregnenolone | – | – | – | 0.35 | 6.00e-03 | – | – |
| N8-Acetylspermidine | – | – | – | 0.30 | 2.00e-03 | – | – |
| Metabolite | Author-emphasized biomarkers | Cutoff value | AUROC (95%CI) | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|---|---|
| creatine | – | – | 0.941 (Creatine + myoinositol + LPE 16:0 ) | 0.844 (Creatine + myoinositol + LPE 16:0 ) | 0.944 (Creatine + myoinositol + LPE 16:0 ) | – |
| 6-phosphonoglucono-D-lactone | – | – | – | – | – | – |
| 2-hydroxy-6-aminopurine | – | – | – | – | – | – |
| 8-hydroxy-2-deoxyguanosine | – | – | – | – | – | – |
| guanosine | – | – | – | – | – | – |
| myo-inositol | – | – | 0.941 (Creatine + myoinositol + LPE 16:0 ) | 0.844 (Creatine + myoinositol + LPE 16:0 ) | 0.944 (Creatine + myoinositol + LPE 16:0 ) | – |
| Cer(d18:0/16:0)-2 | – | – | – | – | – | – |
| LPC 15:0 sn-1 | – | – | – | – | – | – |
| LPC 17:0 sn-1 | – | – | – | – | – | – |
| LPE 16:0 sn-1 | – | – | 0.941 (Creatine + myoinositol + LPE 16:0 ) | 0.844 (Creatine + myoinositol + LPE 16:0 ) | 0.944 (Creatine + myoinositol + LPE 16:0 ) | – |
| LPE 18:1 sn-1 | – | – | – | – | – | – |
| PC 36:0 | – | – | – | – | – | – |
| PC 38:1 | – | – | – | – | – | – |
| PC O 36:1 | – | – | – | – | – | – |
| SM 35:2 | – | – | – | – | – | – |
| 6-phosphogluconic acid | – | – | – | – | – | – |
| citric acid | – | – | – | – | – | – |
| lactic acid | – | – | – | – | – | – |
| glyceraldehyde-3-phosphate | – | – | – | – | – | – |
| D-glucose 6-phosphate | – | – | – | – | – | – |
| D-Sedoheptuiose 7-phosphate | – | – | – | – | – | – |
| D-Erythrose 4-phosphate | – | – | – | – | – | – |
| 5-deoxy-5'-methylthioadenosine | – | – | – | – | – | – |
| SAH | – | – | – | – | – | – |
| xanthine | – | – | 0.802 (xanthine + PC 35:2) | – | – | – |
| creatine | – | – | – | – | – | – |
| glutathione | – | – | – | – | – | – |
| oxidized glutathione | – | – | – | – | – | – |
| carnitine C4:0 | – | – | – | – | – | – |
| carnitine C18:1 | – | – | – | – | – | – |
| FFA C20:2 | – | – | – | – | – | – |
| FFA C20:3 | – | – | – | – | – | – |
| Eicosatrienoic acid methyl ester | – | – | – | – | – | – |
| LPC 15:0 sn-1 | – | – | – | – | – | – |
| LPC 15:0 sn-2 | – | – | – | – | – | – |
| LPC 18:1 sn-2 | – | – | – | – | – | – |
| LPC 22:0 sn-2 | – | – | – | – | – | – |
| LPC 22:4 sn-2 | – | – | – | – | – | – |
| LPE 16:1 sn-1 | – | – | – | – | – | – |
| LPE 16:1 sn-2 | – | – | – | – | – | – |
| LPE 18:0 sn-1 | – | – | – | – | – | – |
| LPE 18:1 sn-2 | – | – | – | – | – | – |
| LPE 20:1 sn-1 | – | – | – | – | – | – |
| LPE 22:4 sn-2 | – | – | – | – | – | – |
| LPE 22:6 sn-2 | – | – | – | – | – | – |
| PC 32:0 | – | – | – | – | – | – |
| PC 33:1 | – | – | – | – | – | – |
| PC 33:2 | – | – | – | – | – | – |
| PC 35:2 | – | – | 0.802 (xanthine + PC 35:2) | – | – | – |
| PC 35:3 | – | – | – | – | – | – |
| PC O 34:2-2 | – | – | – | – | – | – |
| PE O 36:3 | – | – | – | – | – | – |
| PE 32:1 | – | – | – | – | – | – |
| PE 38:1 | – | – | – | – | – | – |
| PE 38:4 | – | – | – | – | – | – |
| PE 38:6 | – | – | – | – | – | – |
| PE O 34:3 | – | – | – | – | – | – |
| SM 36:2 | – | – | – | – | – | – |
| sphinganine | – | – | – | – | – | – |
| sphingosine | – | – | – | – | – | – |
| kynurenine | – | – | – | – | – | – |
| oxypurinol | – | – | – | – | – | – |
| pregnenolone | – | – | – | – | – | – |
| N8-Acetylspermidine | – | – | – | – | – | – |
Paper graphical summary